The early hopes of those who have promoted pharmacogenetics and pharmacogenomics as tools for determining the therapeutic use of medications in the individual patient were that gene variants (polymorphisms) would predict which individuals would benefit and which might suffer harms. A third aspiration, linked to the first, was that such variants could also be used routinely to predict effective dosage regimens.
The early hopes of those who have promoted pharmacogenetics and pharmacogenomics as tools for determining the therapeutic use of medications in the individual patient were that gene variants (polymorphisms) would predict which individuals would benefit and which might suffer harms. A third aspiration, linked to the first, was that such variants could also be used routinely to predict effective dosage regimens.
The last of these hopes has not been fulfilled. The most extensively studied drug has been warfarin, but even using two gene variants, based on the main enzyme by which warfarin is metabolized, CYP2C9, and its chief pharmacological target, vitamin K epoxide reductase (VKOR), plus age and sex as covariates, at best the effective dosage in an individual can be predicted with only about 50% probability. Other variants have been added, and according to the Clinical Pharmacogenetics Implementation Consortium (CPIC, “an international consortium of individual volunteers and a small dedicated staff who are interested in facilitating use of pharmacogenetic tests for patient care”) “incorporation of genetic information has the potential to shorten the time to attain stable INR, increase the time within the therapeutic INR range, and reduce underdosing or overdosing during the initial treatment period.”
Nevertheless, the usefulness of genetic testing for initial warfarin dosing is still debatable. It is not a routine part of care in the UK, and INR testing is still necessary during long term treatment, because of non-genetic factors, such as varying liver function and drug-drug or drug-food interactions. Hence current interest in novel oral anticoagulants (NOACs) that do not act via mechanisms involving vitamin K.
As far as benefits are concerned, progress has been made in determining which variants in the genome of a tumour (which is not the same as the host’s genome, a fact that is not generally as well appreciated as it should be) can predict responsiveness to drug therapy. This has progressed to the extent that there are now some compounds under development that may be useful no matter what organ is affected, provided the tumour has the relevant variant.
Predicting adverse drug reactions has been less successful. I cannot recall hearing a lecture on pharmacogenetics in which abacavir was not mentioned. That is because it is the great success story in this area. The risk of serious rashes in those who are given abacavir is closely related, virtually 100%, to the presence of a polymorphism in an HLA gene, HLA B* 57:01. Pretreatment testing is routine and anyone with the variant will not be given the drug.
Other variants have not been introduced so widely. In a review of 261 biomarkers for adverse drug reactions, only 14 (5%) were listed as being covered by all six US insurers, and most of those were for drugs used in cancer treatment; another 14 were listed by any insurer, the non-cancer related compounds being abacavir, allopurinol, carbamazepine, clopidogrel, eliglustat (for Gaucher’s disease), ivacaftor (for cystic fibrosis), and phenytoin. A major problem is the perceived clinical usefulness of predictive genotyping. For example, the polymorphism HLA-B*58:01, which predicts adverse reactions to allopurinol, has a very low frequency in the general population: the number of patients needed to test to prevent one adverse drug reaction is 11 286 , and routine testing would not be cost effective.
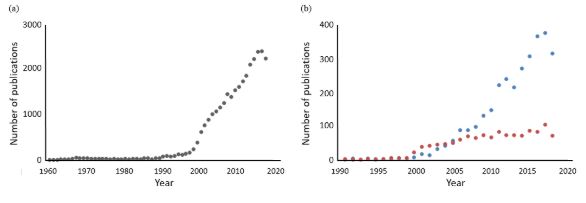
The term “Pharmacogenetik” was introduced by Friedrich Vogel of Heidelberg in 1959, and the earliest paper listed in Pubmed on the subject was a review by David Price Evans and C A Clarke in 1961. It is not surprising that interest in the subject started to burgeon at around the time that the first human genome sequences were published, in February 2001. However, for all the interest in pharmacogenetics/pharmacogenomics, relatively little has been published on the use of biomarkers in predicting the risk of harms and the cost effectiveness of doing so (see Figure 1).
When everyone can come into the doctor’s surgery with their genome listed on their smartphones, some of this may become reality, but the importance of non-genetic factors in drug therapy, and the importance of careful monitoring during therapy, cannot be overstated.
Figure 1. Numbers of hits in Pubmed searches year on year for (a) papers containing terms related to pharmacogenetics and/or pharmacogenomics (PG; total number of hits about 30 000); (b) papers containing terms related to biomarkers connected with PG (blue dots; total number of hits about 3000) and papers containing terms related to pharmacoeconomics connected with PG (orange dots; total number of hits about 1200); note the differences in the dates on the horizontal axes and the numbers of hits on the vertical axes; the apparent decline in all of these in 2018 may be due to incomplete indexing at the time of this search in 2019
Jeffrey Aronson is a clinical pharmacologist, working in the Centre for Evidence Based Medicine in Oxford’s Nuffield Department of Primary Care Health Sciences. He is also president emeritus of the British Pharmacological Society.
Competing interests: None declared.