Challenges faced by the testing system in the UK show that a new way of thinking is required, argue Alan McNally and Jonathan Ball
Mass diagnostic testing for infectious disease—whether for surveillance or for targeted screening—is logistically and financially challenging. This is particularly true when disease prevalence is relatively low and when other unrelated infections present with very similar symptoms. In this kind of scenario, as we are seeing for community SARS-CoV-2 testing now, the low rate of positivity means a significant outlay of resources, time, and effort to identify relatively low numbers of cases.
But this challenge isn’t new, nor is the potential solution. Testing pooled samples, rather than each individual one, was used in the 1990s to identify virus specific antibodies in both human and veterinary settings. [1,2] When used in HIV antibody surveillance studies in a low prevalence setting, testing multiple pools, each containing five individual samples, reduced costs by 80%, even with a subsequent confirmatory test to identify seropositive individuals.
It was soon realized that mass testing could also be applied to look for the virus detection. For example, screening of blood donations for the presence of common blood-borne viruses could be performed by pooling multiple serum samples, then testing them directly with PCR tests, albeit with an intermediate concentration step to improve overall sensitivity. [4,5]
Sample pooling continued to be used widely in veterinary diagnostics for both antibody and direct virus detection, but its use in human medicine declined. So, what parameters need to be considered to ensure reliable diagnostic outputs? And how is pooling achieved?
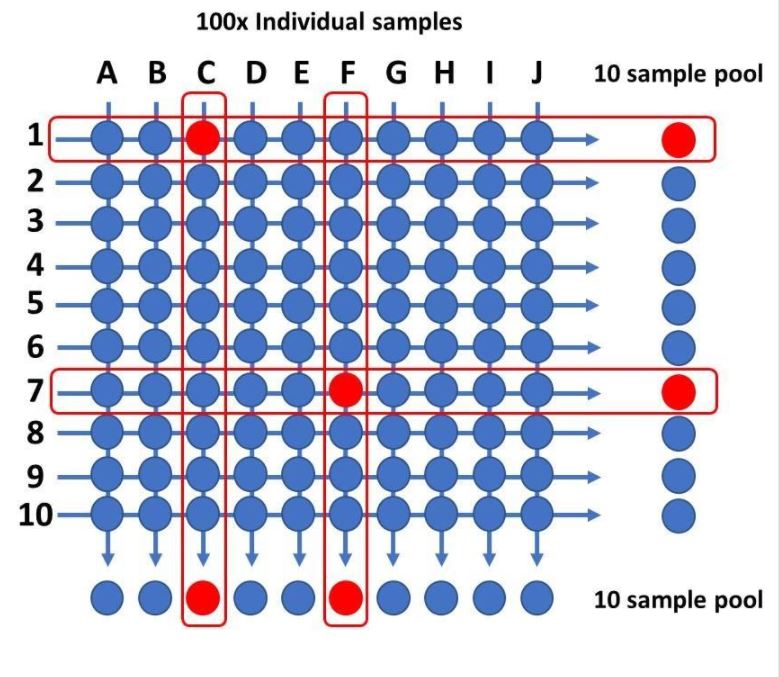
A first key consideration in determining the optimum pooling size is how much test sensitivity decreases as the pool size increases, and this will be determined by the viral load in any positive sample and its prevalence in a background of negative samples. One recent study suggested that pooling up to 30 samples had minimal impact on overall detection rates of SARS-CoV-2, although other have raised concerns that pooling, especially using large pool sizes, can be difficult to deconvolute to identify individual positives. It might also hide technical deficiencies, such as insufficient sampling, and miss low viral load samples. [6,7] One way to tackle the challenges of positive pool deconvolution is to perform matrix pool testing—the most simple being a two-way matrix. [8] In 1-way pool testing, if a pooled sample tests negative, then all samples within that pool are deemed negative. However, if a pool tests positive, then individual samples making up that pool are repeat tested—either in sequentially reducing pool sizes or as individual samples—to identify which were positive. That’s why large pool sizes are more challenging to deconvolute. However, in two-way pooling, a grid or matrix of samples are formed, and column and row pools are generated and tested. In the example shown (see image below), 100 samples are pooled using a 10×10 matrix to produce 20 pools for testing. Only the samples that lie at the intersections of positive columns and rows then need to be tested to identify the positive ones. An additional advantage of pool testing is the increased positive predictive value resulting from confirmatory testing. [9]
The current challenges being faced by the Pillar 2 testing system in the UK show that a new way of thinking is required in our SARS-CoV-2 testing strategy to get back ahead of virus transmission. Much of our testing capacity is taken up by what is essentially asymptomatic screening, whether it is testing of hospital inpatients and staff, testing in care homes, or lack of triage for members of the public arriving at test centres. At the same time there is no surveillance supporting schools, universities, and businesses as they attempt to return to normal.
Pooled sampling of what is essentially asymptomatic swabs would allow rolling surveillance programmes in schools, care homes, universities and colleges, and high-risk factory settings without having a major impact on the daily covid-19 test capacity in the UK. These pooled samples could be tested in labs dedicated to pooled sample testing meaning well-established labs would not need to suddenly change or adapt their current automated protocols. This would allow the UK to improve its ability to monitor high-risk populations and environments, while still having the testing capacity to deal with true symptomatic people. It would also allow testing to be used to follow tracing of cases, something which does not currently drive the test and trace strategy in the UK, but is essential in destroying transmission chains of SARS-CoV-2.
Alan McNally, professor in Microbial Genomics at the University of Birmingham
Competing interests: AMN was seconded to Milton Keynes Lighthouse testing lab for a few months at the start of this year. No further competing interests to declare.
Jonathan Ball, professor of molecular virology at the University of Nottingham.
Competing interests: JKB is involved in an asymptomatic COVID PCR Surveillance project at the University of Nottingham. No further competing interests to declare.
Two way pooled testing. One hundred samples are distributed into two sample pools using a 10×10 two-way grid, each pool containing 10 samples and each sample appearing in two separate pools. In the example (above/below), four pools would yield a positive test result, and confirmatory PCR carried out on the four samples sitting on the intersections of the column and row positive pools would identify samples C1 and F7 as positive and C7 and F1 as negative. At very low prevalence the number of confirmatory tests would be reduced as each two-way array is only likely to produce a maximum of two positive pools and therefore a single intersection and likely positive sample.
References:
1. Tamashiro H, Maskill W, Emmanuel J, Fauquex A, Sato P, Heymann D. Reducing the cost of HIV antibody testing. Lancet. 1993 Jul 10;342(8863):87-90. doi: 10.1016/0140-6736(93)91289-x. PMID: 8100916.
2. Boulard C, Villejoubert C. Use of pooled serum or milk samples for the epidemiological surveillance of bovine hypodermosis. Vet Parasitol. 1991 Jul;39(1-2):171-83. doi: 10.1016/0304-4017(91)90072-4. PMID: 1897116.
3. Kantanen ML, Koskela P, Leinikki P. Unlinked anonymous HIV screening of pregnant women in a low-prevalence population. Scand J Infect Dis. 1996;28(1):3-7. doi: 10.3109/00365549609027141. PMID: 9122629.
4. Schottstedt V, Tuma W, Bünger G, Lefèvre H. PCR for HBV, HCV and HIV-1 experiences and first results from a routine screening programme in a large blood transfusion service. Biologicals. 1998 Jun;26(2):101-4. doi: 10.1006/biol.1998.0144. PMID: 9811513.
5. Roth WK, Weber M, Seifried E. Feasibility and efficacy of routine PCR screening of blood donations for hepatitis C virus, hepatitis B virus, and HIV-1 in a blood-bank setting. Lancet. 1999 Jan 30;353(9150):359-63. doi: 10.1016/S0140-6736(98)06318-1. PMID: 9950441.
6. Lohse S, Pfuhl T, Berkó-Göttel B, Rissland J, Geißler T, Gärtner B, Becker SL, Schneitler S, Smola S. Pooling of samples for testing for SARS-CoV-2 in asymptomatic people. Lancet Infect Dis. 2020 Apr 28:S1473-3099(20)30362-5. doi: 10.1016/S1473-3099(20)30362-5. Epub ahead of print. PMID: 32530425; PMCID: PMC7194818.
7. Mishra B, Behera B, Mohanty M, Ravindra A, Ranjan J. Challenges and issues of SARS-CoV-2 pool testing. Lancet Infect Dis. 2020 Jul 14:S1473-3099(20)30463-1. doi: 10.1016/S1473-3099(20)30463-1. Epub ahead of print. PMID: 32679083.
8. Phatarfod RM, Sudbury A. The use of a square array scheme in blood testing. Stat Med. 1994 Nov 30;13(22):2337-43. doi: 10.1002/sim.4780132205. PMID: 7855467.
9. Bilder CR, Tebbs JM. Pooled-testing procedures for screening high volume clinical specimens in heterogeneous populations. Stat Med. 2012 Nov 30;31(27):3261-8. doi: 10.1002/sim.5334. Epub 2012 Mar 13. PMID: 22415972; PMCID: PMC3500568.