By Anthony Isles
——————————————————————————————————————-
I have been a preclinical researcher for 25 years, and in that time, there has been a great deal of change in the way we do and report our science. The direction of change is generally for the better, as researchers and policy-makers have pushed to make things more open and transparent, and to improve the robustness of the science we undertake. However, implementing change is slow and there are often specific barriers for researchers in different disciplines.
Mirroring the science community, my own group is striving to improve our research practice. Here in Cardiff, I work alongside some world-leaders in psychiatric genetics, and the field of genomics has very much grown over the past few decades with the idea of openness, collaboration and data sharing at its core. Consequently, the prospect of making our raw data files, detailed methodology, and analyses easily and freely accessible does not seem that alien to me. Furthermore, this has become easier over the years as more and more platforms that enable data-sharing have come into existence. However, my group is now looking to make our next step towards improving the robustness of our research, as we think about submitting our first Registered report (RR).
Registered Reports
RRs describe a two-stage process to report research findings (Figure 1). Firstly, the study proposal (Introduction and detailed Materials and Methods) are peer reviewed and pre-accepted. Only then can the researchers conduct the study and, provided they adhere to the agreed methods, the research findings, regardless of whether they are positive or negative, will (in principle) be published in full. By focusing the decision about which articles are published on the question and methods, RRs reduce the impact of the many reporting and publication biases that underpin the problems in reproducibility and replicability of research findings1.
These principles seem eminently sensible, so why then are there so few preclinical studies submitted as RRs? One myth is that the reproducibility crisis is a “psychology problem”, that doesn’t apply to the “harder” biological sciences. Although this idea is still persists, following a number of studies and campaigns in the past decade addressing this directly2-4, most preclinical scientists would acknowledge that in actual fact, research practice does need to improve and that the problems of low power, opaque research methodology and dodgy statistics, certainly applies to them too. This lag in acceptance has probably impacted the uptake of RRs by preclinical scientists and there remain many general misconceptions about the RR process, all of which are excellently discussed by Chambers and Tzavella in a recent review1. However, I think that one such issue is a particular barrier to preclinical researchers submitting RRs.
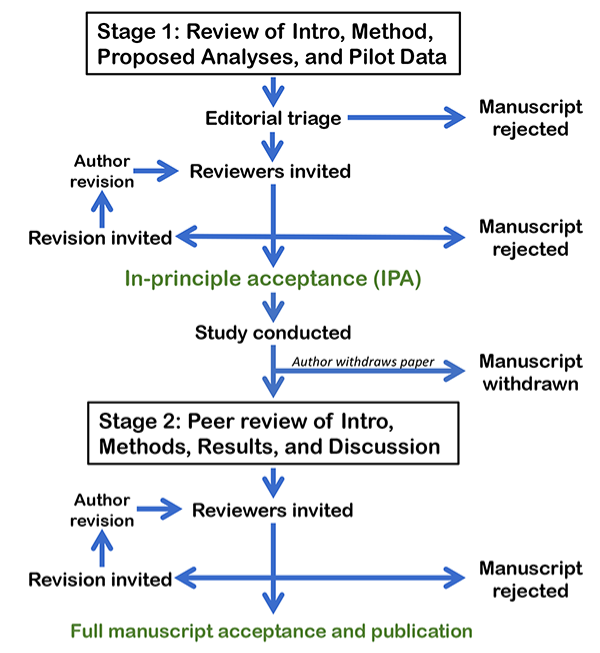
Figure 1 Flow diagram detailing the Registered report process, reproduced with permission from https://cos.io/rr/.
I sometimes joke with my genomics colleagues that they just do one study and then get a number of high-profile papers in “glamour” journals. Sure, these Genome Wide Association Studies (GWAS) involve hundreds of collaborators, collecting thousands of DNA samples from patients and controls, with multiple different analyses of the data, but essentially, they are asking one research question: what is the genetic variation associated with ‘X’? In contrast I would quip, those of us trying to understand the biology of these genetic findings using preclinical models, must undertake a myriad of different experiments and jump through several hoops to get one paper in a regular, jobbing scientific journal.
Joking aside, this is a real issue that has emerged over the last 15 years and I feel is a barrier to the uptake of RRs by preclinical researchers. For example, a key paper from 1984 cited in my PhD thesis was the discovery of genomic imprinting5, 6. The researchers asked one, fundamental question; are the parental genomes of mammals’ equivalent? They tested this hypothesis by generating parthenogenetic and gynogenetic mouse embryos and, when these failed to develop past mid-gestation, concluded that in fact the parental genomes are not functionally equivalent, and there must be an ‘imprint’ that distinguishes them. Essentially, one carefully controlled, pivotal experiment that answered their research question, but also left open many others (no ‘imprinting’ mechanism was described for instance). In contrast, preclinical and basic science papers now involve multiple experiments, using lots of different techniques, figures with >10 panels, and supplementary data that far exceeds the data presented in the main text. This trend has been driven by journal editors and reviewers (and, ultimately, those evaluating science and scientists7) wanting more and more mechanistic details, and this leads to conclusive “story-telling” where each separate set of experiments follow on from those previously. But how does one plan for that? How can a preclinical researcher write an RR with proper methodological planning for a series of experiments, when the outcome of the first isn’t yet known?
In actual fact, many journals that publish RRs accept a certain amount of additional exploratory experimentation that wasn’t included in the initial submission. Nevertheless, we need rein in our expectations of how much constitutes a good preclinical study. I am optimistic that this barrier will be slowly eroded. Once more and more preclinical RRs are submitted, the benefits in terms of robustness will emerge, and this form of publication will also become commonplace. I guess it just needs a few pioneers to stick their head above the parapet… wish me luck!
References
- Chambers CD, Tzavella L. The past, present and future of Registered Reports. Nat Hum Behav 2022; 6(1): 29-42.
- Macleod MR, Lawson McLean A, Kyriakopoulou A, Serghiou S, de Wilde A, Sherratt N et al. Risk of Bias in Reports of In Vivo Research: A Focus for Improvement. PLoS Biol 2015; 13(10): e1002273.
- Button KS, Ioannidis JP, Mokrysz C, Nosek BA, Flint J, Robinson ES et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat Rev Neurosci 2013; 14(5): 365-376.
- https://bnacredibility.org.uk
- McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 1984; 37(1): 179-183.
- Barton SC, Surani MA, Norris ML. Role of paternal and maternal genomes in mouse development. Nature 1984; 311(5984): 374-376.
- Clift J, Cooke A, Isles AR, Dalley JW, Henson RN. Lifting the lid on impact and peer review. Brain Neurosci Adv 2021; 5: 23982128211006574.
——————————————————————————————————————-

Anthony Isles leads a research group investigating the contribution of imprinted genes to brain and behaviour, and their impact on neurodevelopmental and neuropsychiatric disease. He completed his PhD at the University of Cambridge and worked at the Babraham Institute, before moving to Cardiff University, where he is Professor of Molecular and Behavioural Neuroscience.
Potential conflicts of interest declared:
Member of the editorial board or associate editor for Genes Brain and Behavior, Brain and Neuroscience Advances, Philosophical Transactions of the Royal Society (B), and Frontiers in Developmental Epigenetics. Trustee of the British Neuroscience Association (BNA) and member of the BNA ‘Credibility Advisory Board’.