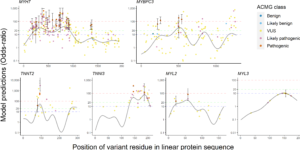
In Mendelian disease genes, missense-variants may cluster in specific functional regions of the protein. Often, there is a complimentary depletion in controls due to population-level constraint. We developed two statistical methods to interrogate this signal, one for gene-association, another for variant interpretation. For gene-discovery efforts, we demonstrate how modeling clustering can improve power. For variant interpretation, we show how parsimonious generalized-additive models are capable of estimating regional burden throughout a linear protein sequence, highlighting mutational hotspots. This proved informative when applied to core hypertrophic cardiomyopathy causing genes with extension to integrate pathogenicity prediction scores. An associated R package and web application facilitates the usage of these methods. (By Adam Waring, https://jmg.bmj.com/content/early/2020/07/30/jmedgenet-2020-106922 )
Data-driven modelling of mutational hotspots and in silico predictors in hypertrophic cardiomyopathy
(Visited 109 times, 1 visits today)