Modeling the genetic risk prediction of type 1 diabetes
Matthew Tien1 and Hui-Qi Qu2
1 University of Texas at Austin, School of Biological Sciences;
2 University of Texas Health Science Center Houston, School of Public Health
Genome-Wide Association Studies (GWAS) perform genome-wide analysis of individuals to screen for single nucleotide polymorphisms (SNP); such studies conducted in a population can detect genetic markers for gene-associated diseases, e.g. type I diabetes (T1D) or Crohn’s disease. With large population GWAS data sets, it is possible to train the data from these studies to develop disease prediction models for clinical purposes. Mathematical models such as logistical regression (LR) and Support Vector Machines (SVM) are able to quantify genotype data from these GWAS and aid the prediction and diagnosis of diseases in individuals. In a recent T1D paper(Wei, Wang et al. 2009), Wei et al. used a larger population of gene markers and used a SVM model to predict the risk of T1D.
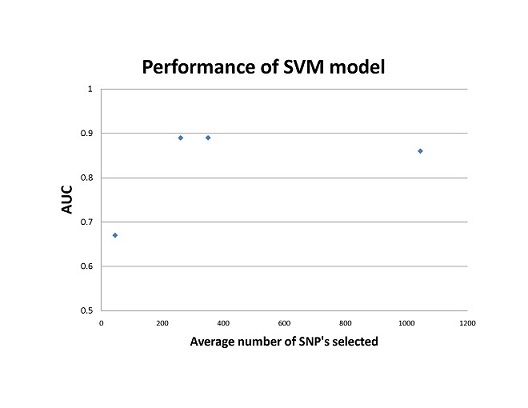
Compared to previous models based on small a small population of 45 markers, SVM and LR models trained with more genetic markers increased the performance of these models; however, using too many markers may include falsely associated markers, thus decrease the accuracy of the model as shown in Fig.1. The SVM model out-competed the LR model in cross-validation testing, area-under-the-curve tests, and multiple sensitivity tests. The SVM model is able to consider multiple genetic interactions of the loci by implementing different kernel functions, something that large multiple LR models cannot calculate. The downfall of SVM modeling however, is the lack of biological interpretability of the transformation of genotypic data.
Figure 1. SVM performance based on the study by Wei et al. The AUC(area under receiver operating characteristic curve) scores are plotted against the mean number of SNP’s selected for each model. The mean was calculated from the range of SNP’s used to train each SVM model.
Use of these models outside of T1D genetic risk determination is limited to diseases with highly genetic susceptibility. These models may be further improved by taking in environmental factors. With more studies, there is significant promise to train such models to accurately predict the genetic risk of developing a heritable disease or condition.
* Matthew Tien is a student at the University of Texas at Austin who is interested mathematical modeling of biological systems, bioinformatics, genomics, and proteomics. He is working with Hui-Qi Qu on the prediction modeling of genetic risk of human complex diseases.
Reference:
Wei, Z., K. Wang, et al. (2009). “From disease association to risk assessment: an optimistic view from genome-wide association studies on type 1 diabetes.” PLoS Genet 5(10): e1000678.