Professor El-Omar has selected Dr Victoria Gudiño PhD and Dr Azucena Salas PhD from Inflammatory Bowel Disease Unit, Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), Hospital Clinic of Barcelona, Barcelona, Spain and Dr Raquel Bartolomé-Casado PhD from the Department of Molecular Oncology, Institute for Cancer Research, Oslo University Hospital-Radiumhospitalet, Oslo, Norway, to do the next #GUTBlog.
The #GUTBlog focusses on the ‘Recent Advances in Basic Science paper “Single-cell omics in inflammatory bowel disease: recent insights and future clinical applications” which was published in paper copy in GUT in August 2025.

“Today, over 7 million patients suffer from inflammatory bowel diseases (IBD), which is comprised of ulcerative colitis and Crohn’s disease. Onset often occurs at a young age and frequently lasts a lifetime. Both diseases are characterised by abdominal pain, diarrhea, bleeding, and pronounced fatigue, involving complex interactions among various intestinal cell types, including immune, epithelial, and stromal cells. The mechanisms underlying IBD are not fully understood, yet they often stem from an inappropriate immune response to microbial or self-antigens in genetically predisposed individuals. Traditional research methods, such as bulk transcriptomics, have limitations in providing detailed insights because they average signals across diverse cell types. This restricts the understanding of cellular diversity and complexity within the highly organised intestinal tissue.
Numerous researchers worldwide are now studying IBD by applying single-cell omics, including not only scRNA-seq but also scATAC-seq, scTCR-seq, or scBRC-seq to intestinal cells, which provide expression data at the single-cell level from several thousand cells/sample. These technologies have revolutionised the study of IBD by providing insights on an individual cell level. In particular, single-cell methodologies enable the identification of novel and sometimes rare cell types and provide insights into their potential functions, thereby linking them to disease activity and complications. For instance, by applying these innovations to the study of certain IBD-associated complications, including fibrostenosis, perianal fistulae, complications of the pouch, and creeping fat accumulation, researchers have uncovered novel insights into the mechanisms that can drive their development. Ultimately, this knowledge can support the identification of previously unrecognized therapeutic targets for these hard-to-treat complications.
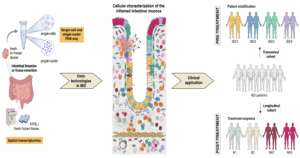
Besides single-cell omic approaches in cell suspensions, emerging techniques, such as spatial transcriptomics and proteomics, are quickly evolving and will no doubt increase our understanding of cellular interactions and environments in situ, showcasing the potential for translational medicine applications. Although still in its infancy, this field is anticipated to enrich the data available for understanding how cells interact within their native contexts, while exploiting the use of old bio-banked fixed tissue samples.
Social media